Reproducible Computational Biology Workflow by nf-core - [Proj:My Computer Workbench]
Introduction
For this part of My Computer Workbench, I would like to introduce about nf-core and their benefits in computational biology.
nf-core
I have also used conda, but setting dependencies can be quite complex, especially on HPC where you are not the administrator. Most of the time, I had to install programs and create the reference genome index I needed by myself.
Bugs started occurring since then. For these reasons, I decided to use a docker container for heavy analysis, such as the genome alignment. One of the convenient ways to do this is by using nf-core.

The nf-core provides analysis pipelines for bioinformatics. It is a workflow run by Nextflow and includes the necessary programs. For example, RNA-seq analysis, where I can select which program I will use for each process. Currently, I am running the upstream process with nf-core on an HPC.
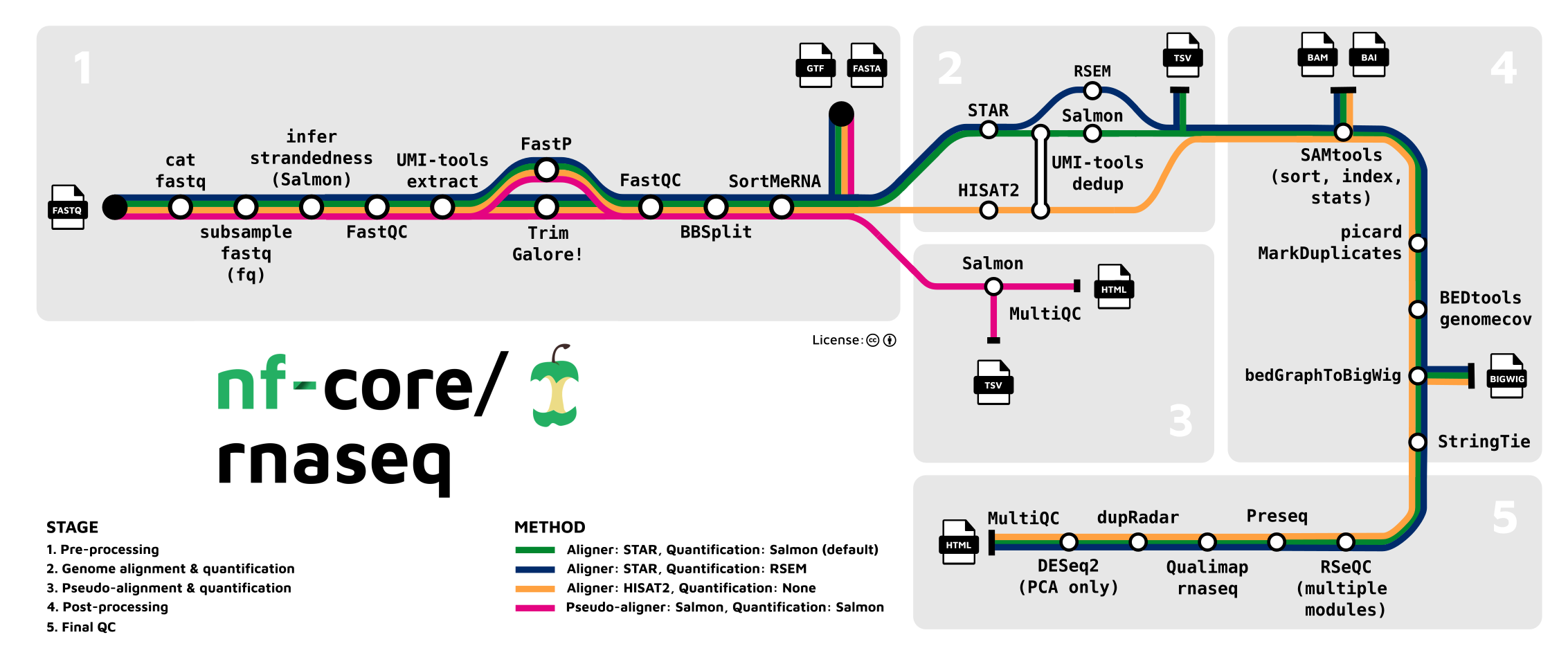
To use nf-core pipelines, I installed it as recommended in the Getting started. Roughly, there are 3 main steps I did on the HPC, including nextflow installation, docker installation (you can choose others such as conda), and aliasing nf-core.
For the alias step, I used the following command. This tells the HPC that whenever I type nf-core means “docker run -itv pwd:pwd -w pwd nfcore/tools”. Please note that there is a difference between nfcore and nf-core in the command.
alias nf-core="docker run -itv `pwd`:`pwd` -w `pwd` nfcore/tools"
To test if the alias works, use the following command. This command will list all nf-core pipelines.
nf-core list