Popular Computational Biology Language with R - [Proj:My Computer Workbench]
Introduction
For this part of My Computer Workbench, I would like to explain the programming language I am using. Additionally, this section covers trick for R and how to use R with reproducibility in mind.
R and R studio
I mostly use R and Rstudio mostly for my downstream analysis and visualization. However, I have encountered program dependency problems when running R on Linux. As a solution, I have started using dockerized RStudio to create an RStudio server.
Here is the code for running dockerized RStudio. Step 1 Create directory for the Dockerized R studio
cd /home/namthip
mkdir R
cd R
mkdir host-site-library
Step 2 Give writable permission on host-site-library, to install custom R packages
chmod a+w -R /home/namthip/R/host-site-library
Step 3 To initialize the Dockerized R Studio and open RStudio Server
sudo docker run \
-v /home/namthip:/home/rstudio/namthip \
-v /home/namthip/R/host-site-library:/usr/local/lib/R/host-site-library \
-e R_LIBS_USER=/home/namthip/R/host-site-library \
-e PASSWORD=<MY_PASSWORD> \
-p 8787:8787 \
bioconductor/bioconductor_docker:RELEASE_3_15
I pull a Docker image from bioconductor (as suggested by Tommy in his blog post). I think this Docker image is great. They explain clearly how to mount a volume (directory). I had tried other Docker images and found that some cannot mount the volume, while others cannot install new packages. This is my own RStudio server on my computer.
Please note that the path that you use in the dockerized R is the path in the container, otherwise dockerized R can’t find your destination. You can easily find the path by using Terminal in the dockerized R studio. Going to the directory you want, then use pwd command to find the path of that directory.
Trick
1. R version
One big problem I found with R version issues is the compatibility between the R version and package version. Sometimes we need a newer version, but sometimes the old one works fine. To switch between R versions, I use this trick.
Docker allows us to pull a specific version of a docker image, as shown in the picture (or this link.)
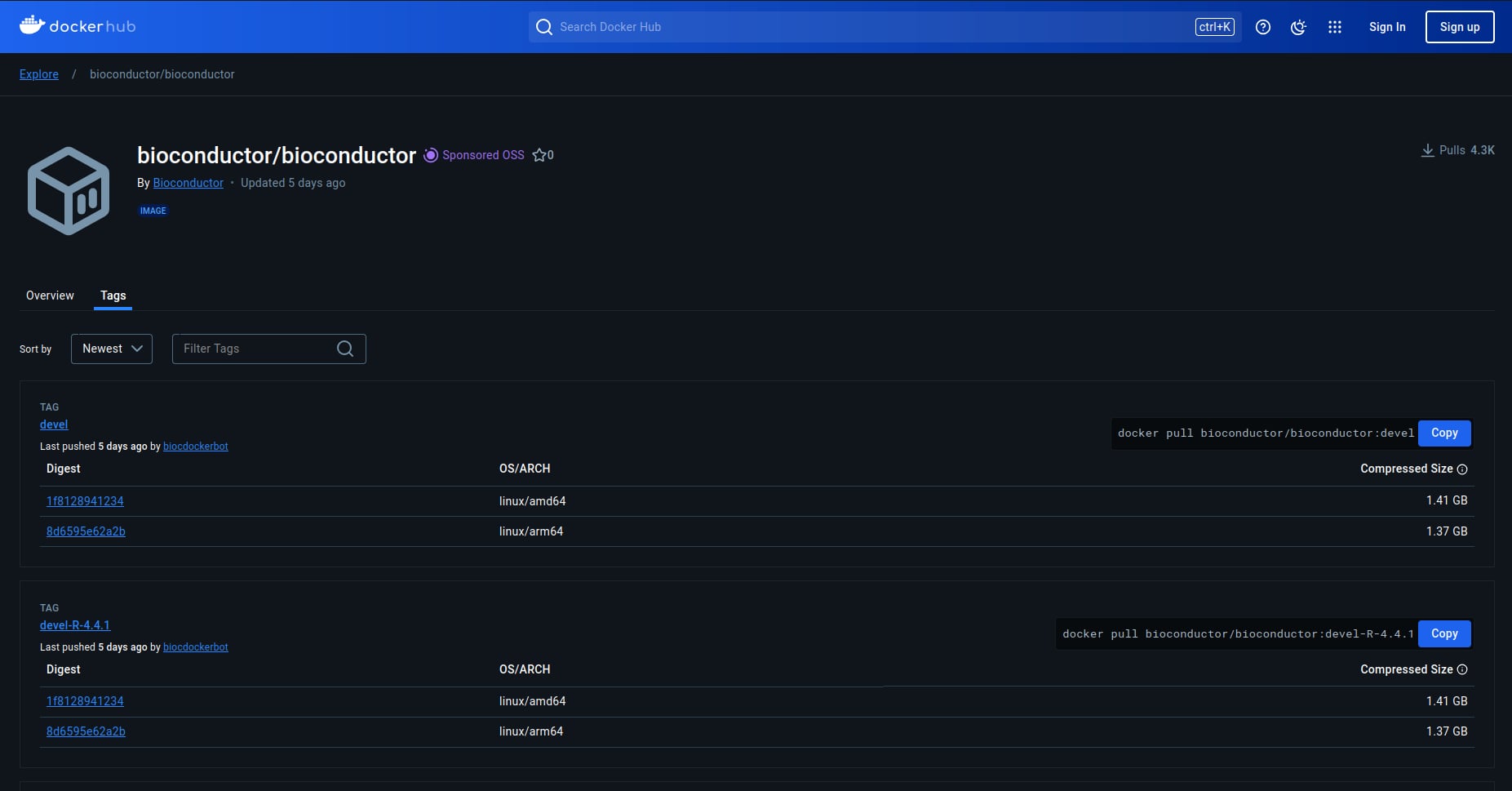
The version of R will be shown in the docker file for each version. From the picture below, “R version 4.4.1” is stated on line number 7.
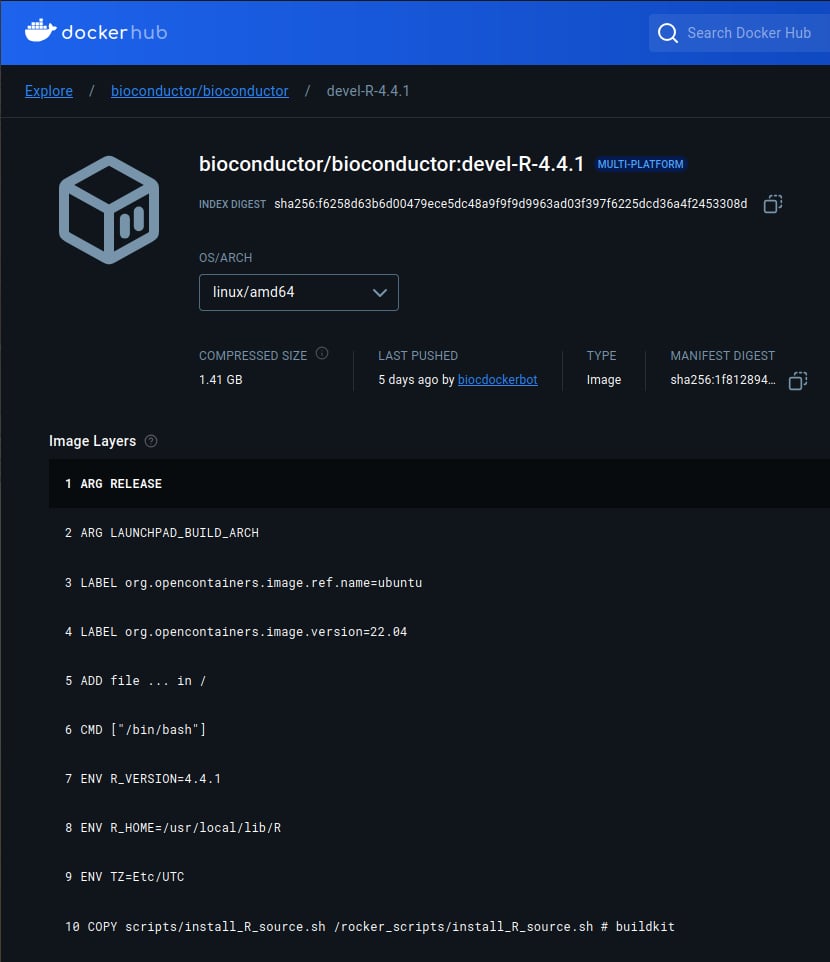
From this point, I pull “R version 4.4.1” from docker, mount the same volume, and initialize a Dockerized R Studio and open RStudio Server using the command line below.
sudo docker run \
-v /home/namthip:/home/rstudio/namthip \
-v /home/namthip/R/host-site-library:/usr/local/lib/R/host-site-library \
-e R_LIBS_USER=/home/namthip/R/host-site-library \
-e PASSWORD=<MY_PASSWORD> \
-p 8787:8787 \
bioconductor/bioconductor_docker:RELEASE_3_19-R-4.4.1
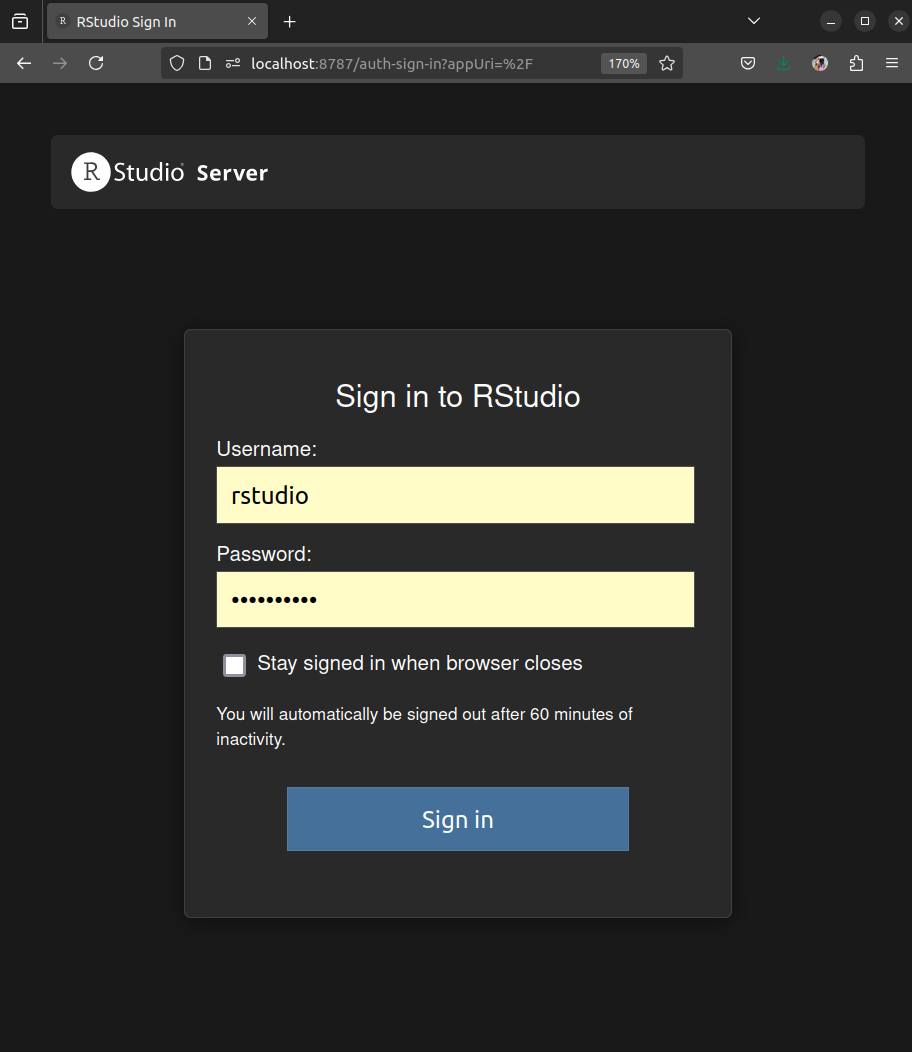
After launching the container, we need to access RStudio Server by opening an internet browser and typing localhost:8787. The login page for RStudio Server will appear. The username will always be rstudio and the password is the one you defined.
2. Best practice for R
If you start using R, it is good to do so following this suggestion “Best Practice for R”. I started using R quite from scratch; I had to fix so many things afterward.
