What are GO, MSigDB, KEGG, ORA, and GSEA?
What are those terms in transcriptomics analysis?
Let’s start with the full name of each
- Gene Ontology (GO)
- Molecular Signatures Database (MSigDB)
- Kyoto Encyclopedia of Genes and Genomes (KEGG)
- Over-Representation Analysis (ORA)
- Gene Set Enrichment Analysis (GSEA)

Picture generated by Gemini, Prompt by Krittiyabhorn Kongtanawanich.
Grouping according to the usage
1. Source of gene set -> GO, msigDB, and KEGG
1.1 Gene Ontology (GO)
- GO provides a structured vocabulary for annotating genes and gene products based on their biological processes, cellular components, and molecular functions. It is widely used to categorize genes into meaningful biological contexts.
- GO includes BP, CC and MF
1.2 Kyoto Encyclopedia of Genes and Genomes (KEGG)
- KEGG is a database that provides information on biological pathways, diseases, and drugs. It includes curated pathways that represent molecular interactions and reactions in various biological systems. KEGG pathways are often used in conjunction with GO for pathway analysis.
- KEGG includes pathway, module, enzyme, network, drug and disease
1.3 Molecular Signatures Database (MSigDB)
MSigDB is a collection of annotated gene sets that can be used for gene set enrichment analyses. It includes gene sets derived from various sources, including GO terms, KEGG pathways, and other curated datasets. Researchers use MSigDB to access predefined gene sets for enrichment analyses.
1.4 Other sources of gene set
Other sources of gene set:
- WikiPathway
- EnrichrDb
- Reactome
- MeSH
- DisGeNET
- Disease Ontology (DO)
- Network of Cancer Gene (NCG) (version 6 and v7)
- The COVID-19 Drug and Gene Set Library.
2. Application in Analyses -> ORA and GSEA
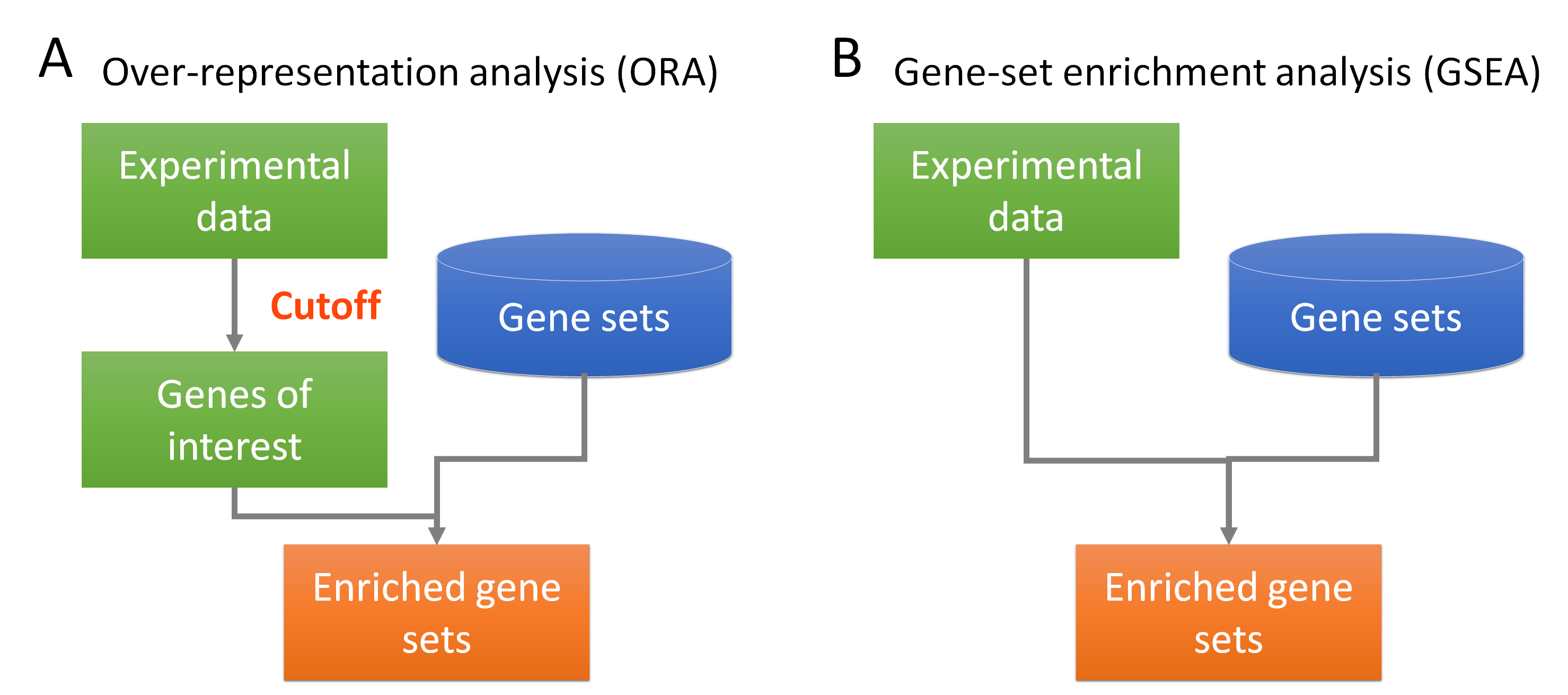
2.1 Over-Representation Analysis (ORA)
- ORA is a statistical method used to determine if a set of genes (e.g., differentially expressed genes) contains a higher proportion of genes associated with specific biological functions or pathways than would be expected by chance. This method often uses databases like GO and KEGG to assess enrichment.
- In ORA, researchers might input a list of differentially expressed genes to see if they are over-represented in specific GO terms or KEGG pathways.
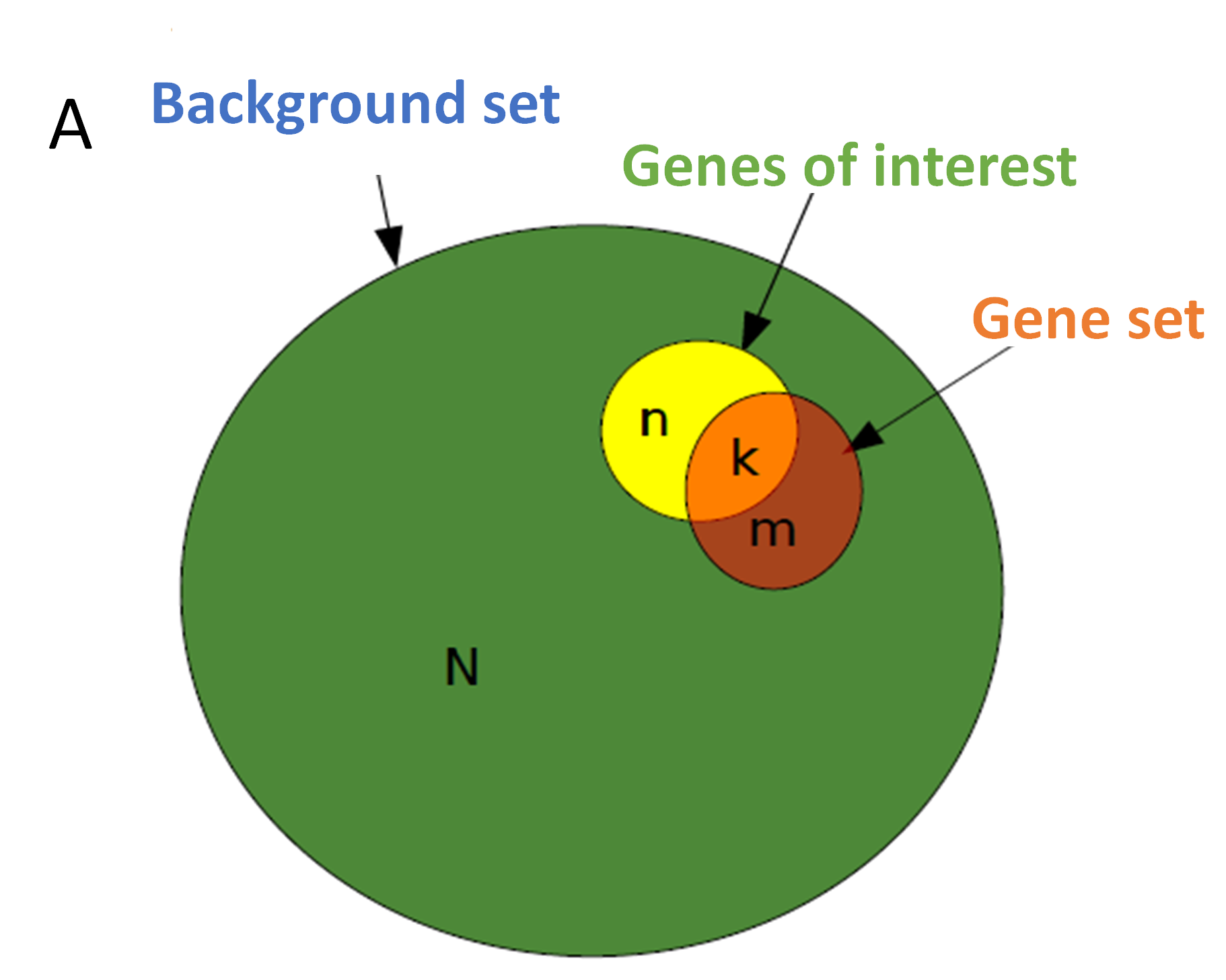
How the genes in ORA were defined. Credit pic
2.2 Gene Set Enrichment Analysis (GSEA)
- GSEA evaluates whether predefined sets of genes show statistically significant differences between two biological states (e.g., treated vs. untreated). Unlike ORA, GSEA considers all genes in the dataset and ranks them based on expression levels, allowing it to detect subtle changes across entire gene sets.
- In GSEA, the entire list of ranked genes can be assessed against the same or similar sets from msigDB, allowing for a broader understanding of the biological implications of expression changes.
- For more information regarding GSEA issue, please read a post from Ming (Tommy) Tang.
Overview of GSEA. Credit pic
Summary of the connections
In summary, GO provides functional annotations, KEGG offers pathway information, msigDB aggregates these resources into usable gene sets, while ORA and GSEA are methodologies that leverage these databases to interpret gene expression data in a biological context.

The different between ORA and GSEA. Credit pic