CCA Stem Cell
Cholangiocarcinoma Stem Cell Research
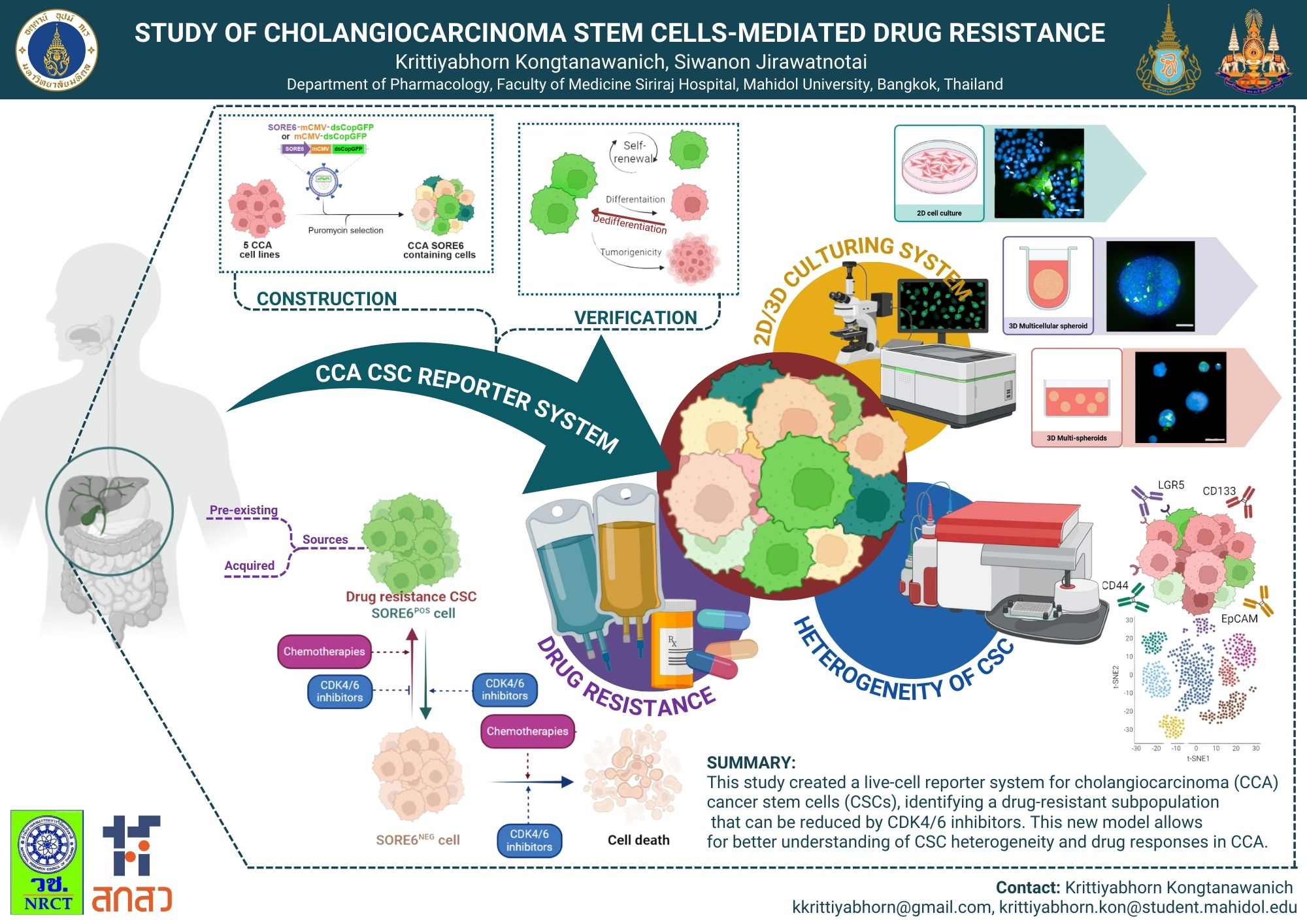
Cholangiocarcinoma (CCA), a malignancy with a dismal prognosis, is characterized by a high proportion of cancer stem cells (CSCs). Traditional methods for identifying CCA CSCs often fail to capture their dynamic nature. This study demonstrated a correlation between poor patient outcomes and the dual expression of SOX2/OCT4. Consequently, we established CCA cell lines harboring a live-cell CSC-specific reporter (SORE6) to track these cells. Analysis of five CCA cell lines revealed SORE6-positive (SORE6+) cells. These CCA CSC models were validated by assessing key CSC properties, including self-renewal, differentiation potential, and tumorigenicity. Furthermore, the reporter system, in conjunction with cell surface markers, allowed for the characterization of CCA CSC heterogeneity. Under standard CCA chemotherapies, the SORE6+ CSC population was enriched, likely due to intrinsic drug resistance and dedifferentiation capabilities. Importantly, treatment with CDK4/6 inhibitors was found to reduce the SORE6+ CCA cell population.
🔬 CSC Models Developed
In addition to conventional assays, we developed and validated three complementary CSC models to better represent tumor biology:
- 2D SORE6 Reporter System – a live-cell biosensor for real-time CSC tracking in CCA cell lines.
- 3D Multicellular Spheroid Model – currently a complex 3D cell architecture model that better mimics tumor-like structure. While not yet a full co-culture, it has potential to be extended toward stromal and immune interactions in future work.
- 3D Multi-spheroids Model – a high-throughput culture that enables quantification of tumorigenic potential and drug response at scale.
The 3D multi-spheroids model formed the foundation of our recent publication in Scientific Reports (2025) (Kongtanawanich et al., 2025), where we integrated high-content confocal imaging and computational pipelines for intensive quantitative CSC analysis and drug-response profiling.
📚 Publications
This project has yielded significant results, including:
- Original research articles
- Kongtanawanich K, Jamnongsong S, Hokland M, Wattanapanitch M, Jirawatnotai S. High-content confocal analysis of tumorigenesis, cancer stem cells, and drug response in 3D cholangiocarcinoma cultures. Sci Rep. 2025;15(1):31387. (Kongtanawanich et al., 2025)
- Kongtanawanich K, Likasitwanakul P, Wattanapanitch M, Jirawatnotai S. Optimization of in vitro cell culture conditions suitable for the cholangiocarcinoma stem cell study. Sci Asia. 2025;51(4):1-8. (Kongtanawanich et al., 2025)
- Kongtanawanich K, Prasopporn S, Jamnongsong S, Thongsin N, Payungwong T, Okada S, et al. A live single-cell reporter system reveals drug-induced plasticity of a cancer stem cell-like population in cholangiocarcinoma. Sci Rep. 2024;14(1):22619. (Kongtanawanich et al., 2024)
- Presentations at both international and national conferences:
- 45th Thai Pharmacological and Therapeutic Society Conference: Poster presentation on *“Analyzing Anchorage-Independent Tumorigenesis of the Live Cholangiocarcinoma Stem Cell Biosensor Using High-Throughput Imaging and Computational Pipelines” (Kongtanawanich et al., 2024).
- The Cholangiocarcinoma Foundation’s Annual Conference 2022: Poster presentation on “Live-biosensor identified subpopulations of cholangiocarcinoma cell lines that contain cancer stem cell properties” (Kongtanawanich et al., 2022).
- 42nd Thai Pharmacological and Therapeutic Society Conference: Poster presentation on “Differentiation potential of cholangiocarcinoma stem cells evaluated by live-biosensor” and awarded Best Poster Presentation (Kongtanawanich et al., 2021).
🔗 Related Projects and Resources
-
3D-SiSP for 3D multi-spheroid quantitative analysis Project page →
-
Computational Biology Portfolio
- Phenomics – Coding × High-Throughput Imaging (2D/3D Models) Project page →
- The 3D multi-spheroid model was central to both (Kongtanawanich et al., 2024), (Kongtanawanich et al., 2025) and (Kongtanawanich et al., 2025), with intensive quantitative analysis showcased in the latter.
- This project also empowers wet-lab data from three models (2D, 3D multi-spheroid, and 3D multicellular spheroid) by integrating dry-lab coding pipelines to generate deeper insights into cancer biology.
- Phenomics – Coding × Flow Cytometry Project page →
- Focused on exploring the heterogeneity of CSCs by identifying subpopulations of CCA CSCs.
- Wet-lab flow cytometry data are empowered by dry-lab coding, enabling intensive exploration of CSC diversity and drug resistance mechanisms.
- Phenomics – Coding × High-Throughput Imaging (2D/3D Models) Project page →
This integrated framework of wet-lab innovation and computational biology pipelines supports both mechanistic discovery in CSC biology and the translational development of new therapeutic strategies for cholangiocarcinoma.
References
2025
2024
-
 Analyzing Anchorage-Independent Tumorigenesis of the Live Cholangiocarcinoma Stem Cell Biosensor Using High-Throughput Imaging and Computational PipelinesMay 2024
Analyzing Anchorage-Independent Tumorigenesis of the Live Cholangiocarcinoma Stem Cell Biosensor Using High-Throughput Imaging and Computational PipelinesMay 2024
2022
-
 Live-biosensor identified subpopulations of cholangiocarcinoma cell lines that contain cancer stem cell propertiesFeb 2022
Live-biosensor identified subpopulations of cholangiocarcinoma cell lines that contain cancer stem cell propertiesFeb 2022
2021
-
 Differentiation potential of cholangiocarcinoma stem cells evaluated by live-biosensorFeb 2021
Differentiation potential of cholangiocarcinoma stem cells evaluated by live-biosensorFeb 2021