Genomics (SNP)
My genomics (SNP analysis) portfolio
Introduction
My passion for disease biology has led me to systematically investigate the multilayered mechanisms of gene regulation. I’ve already established a deep understanding of the regulatory landscape through my prior work:
🎨 Epigenomics (ChIP-seq): Analyzing the dynamic blueprint of the epigenome (see my Epigenomic portfolio).
🎨 Transcriptomics: Investigating the functional output of gene expression (see my Transcriptomics portfolio).
However, a fundamental piece of the puzzle has been missing: Genomics. To achieve a truly holistic view—moving beyond how the code is regulated to encompass the primary variations within the code itself—I must delve into the DNA sequence.
This portfolio marks my entry into the essential world of genomics, focusing specifically on Single Nucleotide Polymorphism (SNP) analysis. This foundational work is crucial for identifying the genetic variations that drive disease susceptibility.
I am extremely fortunate to be mentored in this new domain by Dr. Metha Yaikwawong, a respected expert in SNP analysis. Under his guidance, I am committed to bridging the gap between underlying genetic variation, epigenetic control, and resulting transcriptomic profiles, thereby contributing a complete picture to the field of precision medicine.
Overview of Genome Analysis
Firstly, I would like to clarify that ”genome analysis” ≠ only “variant analysis”, but variant analysis (detecting mutations) is one major branch within it. And SNPs analysis is under variant analysis!
1️⃣ What “Genome Analysis” Actually Includes
Genome analysis is a big umbrella — it includes:
| Category | What it studies |
|---|---|
| Structural / Variant analysis | Detects mutations: SNPs, Indels, CNVs, SVs |
| Functional analysis | Studies how genes are expressed or regulated (RNA-seq, methylation, etc.) |
| Comparative / Evolutionary genomics | Compares genomes across individuals or species |
| Epigenomic analysis | Looks at DNA methylation, histone modification |
| Metagenomics | Sequences all DNA from an environment or microbiome |
2️⃣ Variant (Mutation) Analysis — the Core Idea
When people say they “detect mutations” from sequencing data, what they really mean is they find differences between a sample genome and a reference genome. These differences are your “variants” or “mutations”.
🧬 The different between “mutations” and “variants”
| Term | Simple Definition | Neutral or Value-Judgment? |
|---|---|---|
| Variant | Any change in DNA sequence compared to a reference genome | Neutral (just a difference) |
| Mutation | A change in DNA that may alter biological function or cause disease | Value-loaded (often “bad”) |
In modern genomics, people mostly use “variant” because not every DNA change is harmful.
🧬 Terminology in Practice
| Modern Genomics Term | Older Equivalent | Used in |
|---|---|---|
| SNV (Single Nucleotide Variant) | SNP or point mutation | Research / sequencing |
| Pathogenic variant | Disease-causing mutation | Clinical reports |
| Benign variant | Non-disease variant | Clinical reports |
| VUS (Variant of Uncertain Significance) | Unknown mutation effect | Genetic testing |
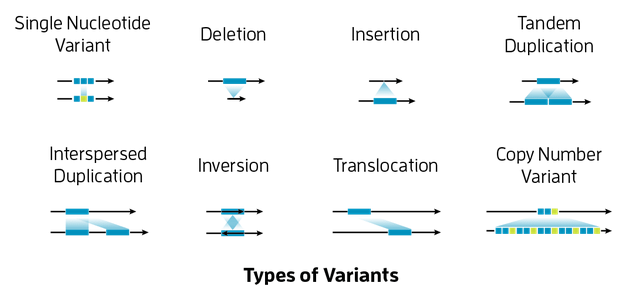
3️⃣ Main Types of Variants
| Variant Type | Description | Example | Size |
|---|---|---|---|
| SNP (Single Nucleotide Polymorphism) | One base changes | A → G | 1 bp |
| Indel (Insertion/Deletion) | Few bases added or removed | +AT or -TG | 1–50 bp |
| CNV (Copy Number Variation) | Gene/region duplicated or deleted | 2 copies → 1 copy | >1 kb |
| SV (Structural Variant) | Rearrangements: inversion, translocation, large insertion/deletion | Chromosome breakpoints | >1 kb |
| Somatic Mutation | Found only in cancer or tissue cells | e.g., KRAS G12D | any size |
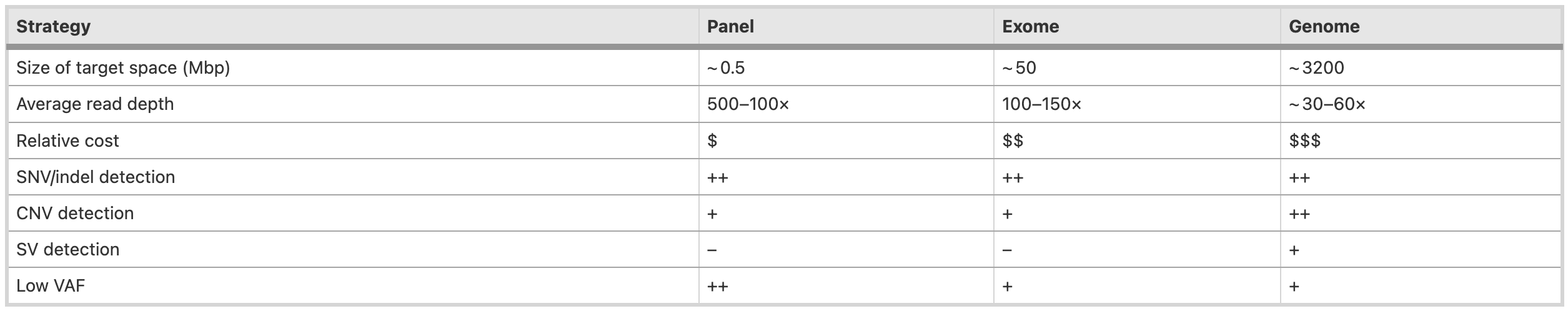
🧬Sequencing strategy for variant calling
4️⃣ Variant Calling Workflow
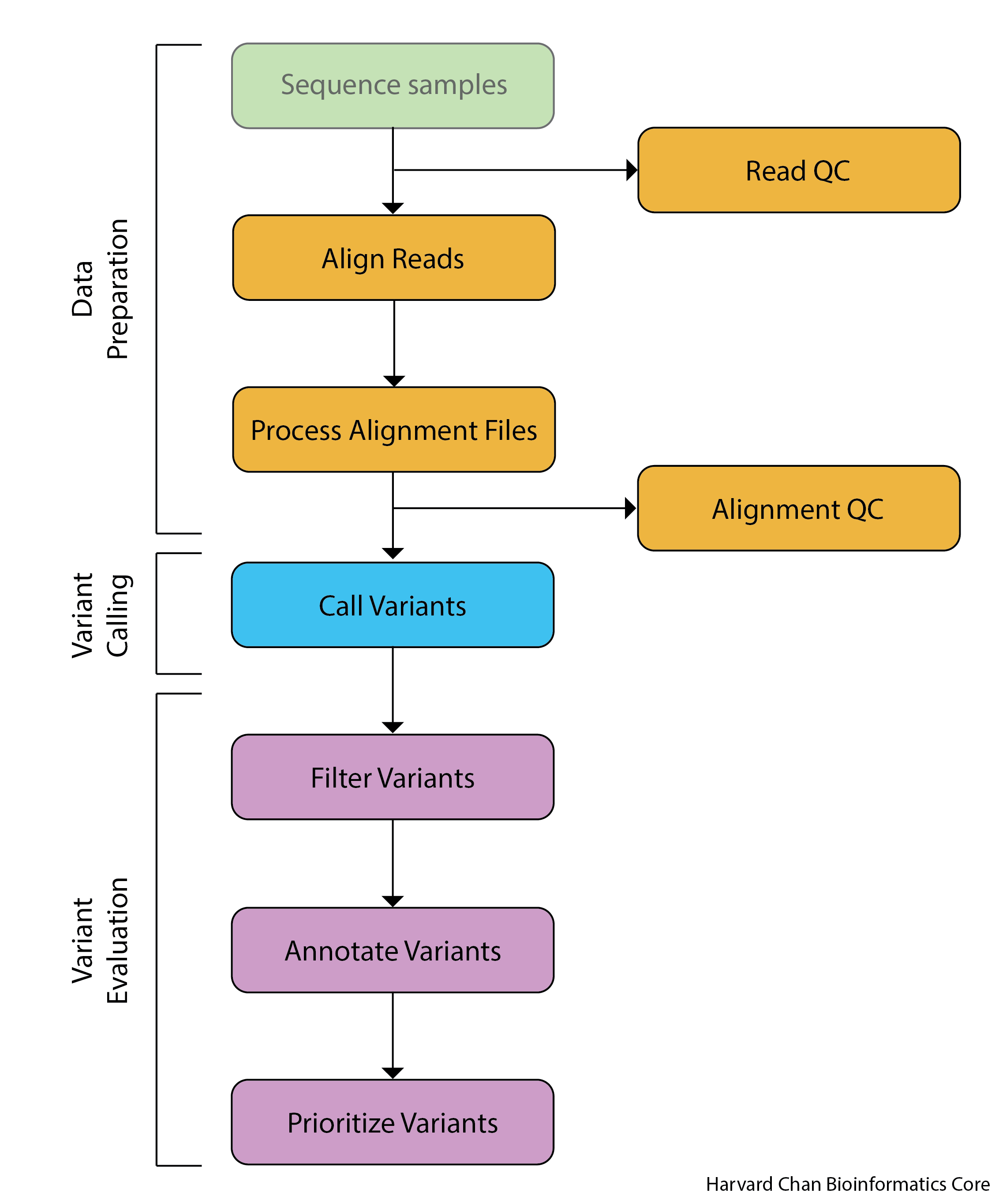
- Sample → DNA Extraction → Sequencing (FASTQ)
🎯Goal: high-quality reads for alignment
🧩Input: paired-end FASTQ files (sample_R1.fastq, sample_R2.fastq)
⚙️Tools: Illumina sequencer, Nanopore, PacBio
↓ - Quality Control (QC)
🎯Goal: Check read quality, adapter contamination, GC content.
⚙️Tools: Adapter trimming (Trimmomatic, Cutadapt),QC report (FastQC, MultiQC)
🧩Output: cleaned FASTQ files
↓ - Alignment to Reference Genome (BAM)
🎯Goal:Map reads to a reference genome (e.g., hg38 for human).
⚙️Tools: BWA-MEM, Bowtie2, HISAT2
🧩Output: SAM → convert to BAM (samtools view -b)
↓ - Post-processing of BAM (Sorting, Duplicate removal, Recalibration)
🎯Goal: Sort reads, mark PCR duplicates, and perform base quality recalibration.
⚙️Tools: samtools sort, Picard MarkDuplicates, GATK BaseRecalibrator
🧩Output: cleaned and indexed BAM file (sample.bam, sample.bai)
↓ - SNP Calling (VCF)
🎯Goal: Compare aligned reads to the reference genome → find positions with base changes.
⚙️Tools: GATK HaplotypeCaller, bcftools mpileup + call, FreeBayes
🧩Output: VCF (Variant Call Format) file \
↓ - Variant Filtering & Quality Control
🎯Goal: Remove false positives and low-quality SNPs.
⚙️Tools: GATK VariantFiltration, vcftools, bcftools filter
🧩Output: high-confidence VCF 🎯Filtering criteria:- Depth (DP) > 10 \
- Quality (QUAL) > 30 \
- Variant allele frequency (VAF) threshold
↓
- SNP Annotation (gene, function, dbSNP ID)
🎯Goal: Add biological meaning to each SNP: gene name, transcript, coding effect, dbSNP ID, clinical significance.
⚙️Tools: ANNOVAR, VEP (Variant Effect Predictor), SnpEff
🎯Databases used:- dbSNP (known SNPs) \
- ClinVar (pathogenicity) \
- gnomAD (population frequency) \
- RefSeq / Ensembl gene models
↓
- Downstream / Biological Interpretation (depends on purpose) \
| Application | Example analysis | Tools |
|---|---|---|
| Population genomics | PCA, Fst, population structure | PLINK, vcftools, ADMIXTURE |
| GWAS | Trait–SNP association | PLINK, GEMMA, SAIGE |
| Phylogenetics / evolution | SNP distance, tree | IQ-TREE, SNPhylo |
| Clinical genomics | Pathogenic variant report | ClinVar, gnomAD, VarSome |
5️⃣ Conceptual Flow
🐚 UNIX (core bioinformatics)
FASTQ ──▶ QC ──▶ Alignment ──▶ BAM ──▶ Variant Calling ──▶ VCF
🧬 The different of Variant Calling (Allele Frequency Expectations): Germline vs Somatic
| Feature | Germline Variant Calling | Somatic Variant Calling |
|---|---|---|
| Genome type | Diploid | Mixed tumor subclones |
| Expected # of alleles | ≤ 2 | Many possible (due to subclones) |
| Typical allele frequency | ~0.5 (heterozygous), 1.0 (homozygous) | Variable (0.05–0.9) |
| Major challenge | Simple genotypes | Distinguish real low-frequency variants from noise |
| Tools | GATK HaplotypeCaller, FreeBayes | Mutect2, VarScan2, Strelka2, LoFreq |
🐍 Python (data science layer)
VCF ──▶ Filtering ──▶ Summary stats ──▶ Visualization ──▶ Report
🧬 Full SNP Analysis Pipeline with Tool Mapping
| Step | Purpose | UNIX / Command Line Tools | Python Libraries / Packages |
|---|---|---|---|
| 1️⃣ Raw data QC | Check sequencing read quality | fastqc, multiqc, trimmomatic, cutadapt | ⚪ (rarely in Python) – can wrap tools using subprocess |
| 2️⃣ Alignment | Map reads to reference genome | bwa mem, bowtie2, hisat2 | ⚪ pysam can read alignment files (BAM/SAM) |
| 3️⃣ SAM → BAM conversion | Compress and sort alignment files | samtools view, samtools sort, samtools index | ✅ pysam (Python bindings for samtools) |
| 4️⃣ Post-processing | Mark duplicates, base recalibration | picard MarkDuplicates, gatk BaseRecalibrator | ⚪ Usually command line (GATK wrapped by Python subprocess if needed) |
| 5️⃣ Variant calling (SNP detection) | Detect SNPs/indels | bcftools mpileup + call, gatk HaplotypeCaller, freebayes | ⚪ Usually done by UNIX (some Python wrappers exist for automation) |
| 6️⃣ Variant filtering | Filter by quality/depth | bcftools filter, vcftools --minQ, grep, awk | ✅ pandas, cyvcf2, PyVCF (for programmatic filtering) |
| 7️⃣ Variant statistics | Ts/Tv ratio, allele frequency | vcftools --TsTv-summary, bcftools stats | ✅ scikit-allel, pandas |
| 8️⃣ Variant annotation | Add gene, effect, dbSNP ID | VEP, ANNOVAR, SnpEff | ⚪ Can parse output in Python for summary |
| 9️⃣ Data visualization | Plot SNP density, allele frequency | — | ✅ matplotlib, seaborn, plotly, scikit-allel |
| 🔟 Population analysis (optional) | PCA, diversity, Fst, GWAS | plink, vcftools, ADMIXTURE | ✅ scikit-allel, numpy, scipy, pandas |
| 1️⃣1️⃣ Reporting | Summarize and export tables | awk, sed, grep, Rscript | ✅ pandas, xlsxwriter, csv, matplotlib |
NOTE
There are so many details about the analysis such as
- Germline and somatic variant calling is different, thus tools and setting are different.
My SNPs analysis
I will start with a VCF files which were finished upstream process.
✅ Good sources of public VCF files Sure! Here’s the same style of writing for ClinVar and other commonly used public VCF sources 👇
✅ Good sources of public VCF files
- ClinVar (NCBI) – Provides curated variant calls with clinical significance (e.g., pathogenic, benign, uncertain significance) for the human genome (GRCh37 & GRCh38). ftp.ncbi.nlm.nih.gov
- gnomAD (Genome Aggregation Database) – Contains population-level allele frequencies for millions of variants across exomes and genomes from diverse human -cohorts. gnomad.broadinstitute.org
- ExAC (Exome Aggregation Consortium) – The predecessor to gnomAD, focused on exome sequencing data from over 60,000 unrelated individuals. gnomad.broadinstitute.org
- 1000 Genomes Project / IGSR – Hosts genome-wide variant data (phase 3) across multiple populations, often used as a standard reference dataset. internationalgenome.org
- Personal Genome Project (PGP) – Contains public genomes of volunteer participants with available VCFs (exome and whole-genome). my.pgp-hms.org
✅ Database as reference catalogue (not provide individual information)
- dbSNP (NCBI) – A comprehensive catalog of all known single nucleotide polymorphisms (SNPs) and short indels reported across studies, organized by chromosome.
1. Data set 1: 1000 Genome
I will download it from 1000 Genome
Steps
- Download file from public resource and cut it to small file for demonstration.
# download
cd /home/kk/Jupyter_docker/SNP_interview/input/1000genome
wget https://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/ALL.chr1.phase3_shapeit2_mvncall_integrated_v5b.20130502.genotypes.vcf.gz
# get only first 500 lines
zcat ALL.chr1.phase3_shapeit2_mvncall_integrated_v5b.20130502.genotypes.vcf.gz | grep '^#' > 1000genome_test.vcf
zcat ALL.chr1.phase3_shapeit2_mvncall_integrated_v5b.20130502.genotypes.vcf.gz | grep -v '^#' | head -n 500 >> 1000genome_test.vcf
# compress to .gz again
bgzip 1000genome_test.vcf
tabix -p vcf 1000genome_test.vcf.gz
# change to .tsv
zcat 1000genome_test.vcf.gz | grep '^#CHROM' | sed 's/^#//' > 1000genome_test.tsv
zcat 1000genome_test.vcf.gz | grep -v '^#' | head -n 500 >> 1000genome_test.tsv
- Inspect the fil: What’s inside a 1000G VCF
Each row (variant record) summarizes one position in the genome and how it varies across individuals.
🧠 Columns (core meaning)
| Column | Example | Meaning | ||
|---|---|---|---|---|
| CHROM | 1 | Chromosome number | ||
| POS | 10506 | Position (1-based) on that chromosome | ||
| ID | rs12345 | Variant ID (dbSNP rsID) | ||
| REF | T | Reference allele from reference genome | ||
| ALT | A | Alternate allele (observed variant) | ||
| QUAL | 100 | Variant quality score (Phred-scaled likelihood) | ||
| FILTER | PASS | Whether the variant passed QC filters | ||
| INFO | AC=5;AF=0.001;AN=5008;NS=2504;DP=8000 | Summary stats for that variant | ||
| FORMAT | GT:DP | Structure of genotype fields in each sample column | ||
| HG00096, HG00097, … | 0 | 1, 0 | 0 | Each person’s genotype (reference/alternate alleles) |
🧠 How to read the INFO field
Here’s what the 1000 Genomes INFO tags mean (Phase 3 standard):
| INFO key | Meaning | Example | Interpretation |
|---|---|---|---|
| AC | Allele Count (number of ALT alleles across all samples) | AC=5 | 5 alternate alleles observed out of all chromosomes sequenced |
| AN | Allele Number (total chromosomes observed) | AN=5008 | For 2504 diploid individuals, total alleles = 2×2504 = 5008 |
| AF | Allele Frequency | AF=0.001 | Frequency = AC/AN = 0.001 → 0.1% |
| NS | Number of Samples with Data | NS=2504 | All samples were genotyped at this site |
| DP | Total Depth (sum of read depth across all samples) | DP=78015 | Combined coverage |
| EAS_AF, EUR_AF, AFR_AF, AMR_AF, SAS_AF | Per-population allele frequencies | EAS_AF=0.0002 | Subpopulation allele frequency |
| VT | Variant Type | VT=SNP | Indicates type: SNP, INDEL, etc. |
| AA | Ancestral Allele | AA=T | Original allele in ancestral genome |
| RS | dbSNP rsID (redundant with ID field sometimes) | RS=12345 | dbSNP ID |
| MQ | Mapping Quality | MQ=60 | Average mapping quality of reads supporting the call |
| QD | Quality by Depth | QD=15 | Normalized quality |
| VQSLOD | Variant Quality Score Log-Odds | VQSLOD=2.5 | Used in recalibration (GATK) |
🧭 Quick biological interpretation of common INFOs
| INFO tag | Low value means | High value means | Clinical relevance |
|---|---|---|---|
| AF | Rare variant | Common polymorphism | Rare = possibly pathogenic (if in ClinVar) |
| QUAL | Unreliable call | Confident call | Filter threshold often >30 |
| DP | Low sequencing coverage | High coverage | Low DP variants may be false |
| FILTER | Failed QC | Passed QC | Use only PASS |
| AC | Found in few alleles | Found in many alleles | Helps compute frequency |
| AN | Poor coverage (missing genotypes) | All samples genotyped | Important for accuracy |
Filtering by Python
- Keep PASS variants only
- Keep SNPs (not INDELs → VT=SNP)
- Keep biallelic sites (len(ALT.split(‘,’)) == 1)
- Apply QC filters (QUAL, DP, etc.)
- Remove common variants if searching for rare disease-causing ones (AF < 0.01)
- Keep variants in coding or regulatory regions
- Annotate with ClinVar, dbNSFP, or custom database
- Filtering according to annotation meaning.
Here is my Jupyter notebook from filtering process 1 to 6.
What I have learned from this data set
This is SNP analysis for an individual person. The key filtering process is rare disease-causing ones (AF < 0.01). Therefore many SNPs still in the panel. We next have to filter by the annotation such as ClinVar. ClinVar will identify whether the SNP are pathogenic, benign, and uncertain significance. Other annotations are also identify diffedrent meaningful SNP such as exon, intron, framship, missense, etc.
Other types of SNP analysis
Compare case (Disease) and control (healthy)
The process:
- Get .VCF from both case and control
- Put the processed data to
PLINKto identify the SNP that different between case and control. - The final analysis result is 2 by 2 table.
🧬 Statistical Analysis in SNP Studies
In SNP (Single Nucleotide Polymorphism) analysis, categorical comparison between groups (e.g., case vs. control) is a key component of statistical testing.
- Comparison Tests
-
Chi-square test — used for categorical data with sufficient expected counts.
-
Fisher’s exact test — applied when any cell in a 2×2 contingency table has an expected count < 5.
-
These tests evaluate the association between genotype (or allele) and phenotype using p-values to determine statistical significance.
-
Predictive Modeling
- Logistic regression — used to assess the association between SNPs and binary outcomes (e.g., disease vs. healthy), adjusting for potential covariates such as age, sex, or population stratification.
-
Effect Size Estimation
- Report odds ratio (OR) and 95% confidence interval (CI) to quantify the strength and direction of the association between the SNP and the trait of interest.
Useful resources
- GWASTutorial by The Laboratory of Complex Trait Genomics at the University of Tokyo
- GWA analysis by Andries Marees
Acknowledgment
- I would like to acknowledge Dr. Metha Yaikwawong for his encouragement and the valuable knowledge he provided on the earlier days of my programming journey.
I’m very happy 🥰 that you are visiting my computational biology portfolio and would be even happier if you could provide suggestions or feedback 🤩.
You can contact me through various online platforms here 📬 or leave a comment below using GitHub account. 👇🏼