Phenomics - Coding x High-Throughput Imaging (2D/3D Models)
My portfolio on high-throughput imaging of 2D and 3D cell cultures and image analysis. :)
Introduction
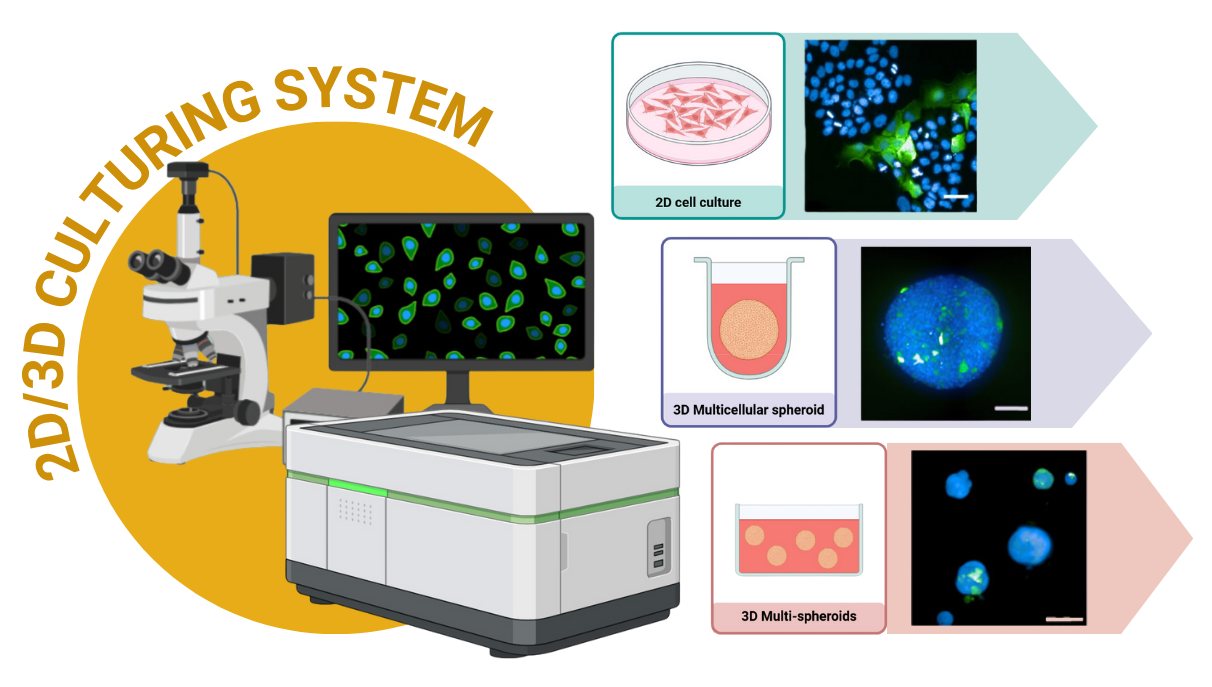
As a proud member of the Siriraj Center of Research Excellence for Systems Pharmacology (SiSP), a leader in high-throughput imaging, I am driven by a passion for uncovering the secrets of cancer biology. My work combines a hands-on approach in the lab with cutting-edge image analysis. I have specialized in exploring a wide array of cellular models, moving from the traditional 2-dimensional (2D) cell cultures to advanced 3-dimensional (3D) multicellular spheroids and multi-spheroid systems. This diverse experience allows me to tackle complex biological questions with creativity and precision. Beyond the visuals, I am also an expert in using RStudio to dive deep into the data, ensuring that every image tells a complete and compelling story.
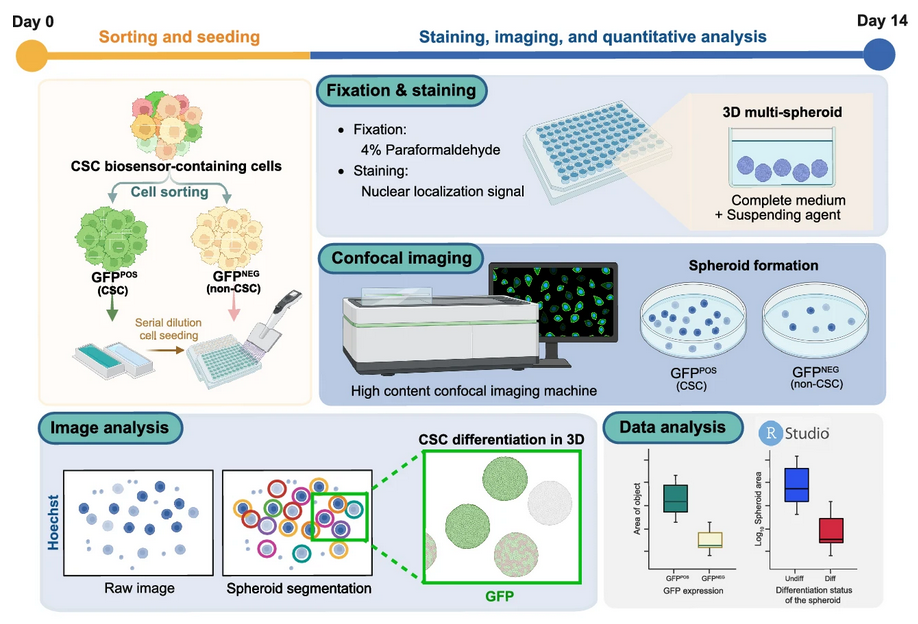
Most recently, this portfolio culminated in a publication in Scientific Reports (2025) (Kongtanawanich et al., 2025), where the 3D multi-spheroid model was used as a core platform for quantitative analysis of tumorigenesis, cancer stem cell (CSC) dynamics, and drug response. This work highlights how wet-lab imaging and computational pipelines can be combined to deliver powerful insights into CSC biology.
Model 2D Cancer Cell Culturing Model
1.1 Analysis overview:
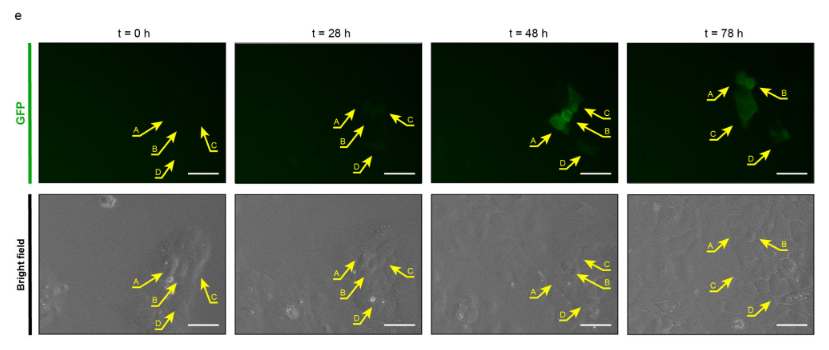
In this model, my work centered on utilizing cancer stem cell (CSC) reporter systems (SORE6 reporter) to study the dynamics of cancer cells in a flat, 2D environment. I employed high-content real-time imaging to precisely monitor the CSC phenotype and understand its behavior over time.
A key part of this work involved a high-throughput drug response study, where I evaluated the effects of various drugs on cancer cells, with a specific focus on their impact on the CSC population. Using this approach, I was able to establish a direct link between CSC biology and drug response. By analyzing the data, I could determine how much a specific drug concentration killed the cells, and in parallel, how that same concentration affected the percentage of cancer stem cells.
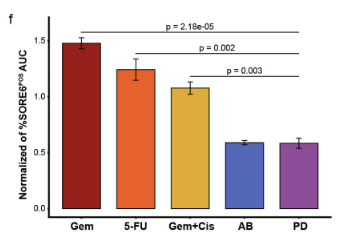
I also developed a detailed step-by-step methodology to create an algorithm that could compare the effects of different drugs on the CSC proportion across a comparable dose range. This systematic approach allowed for a robust comparison of drug efficacies from the perspective of both general cell death and the specific targeting of the CSC population. The imaging data was then analyzed using Columbus™ Image Data Storage and Analysis System (PerkinElmer), enabling a sophisticated, single-cell level analysis. The resulting data was further explored and visualized using RStudio, revealing a comprehensive view of drug efficacy and its influence on CSCs.
1.2 Key Applications:
- Monitoring CSC Phenotype: Used a reporter system with high-content real-time imaging to precisely monitor the behavior of cancer stem cells (CSCs).
-
High-Throughput Drug Screening: Performed drug response studies to evaluate the effects of various drugs on cancer cells, with a specific focus on their impact on the CSC population.
-
Linking CSC Biology and Drug Response: Established a direct link between a drug’s concentration, its cytotoxic effect, and its parallel impact on the percentage of CSCs.
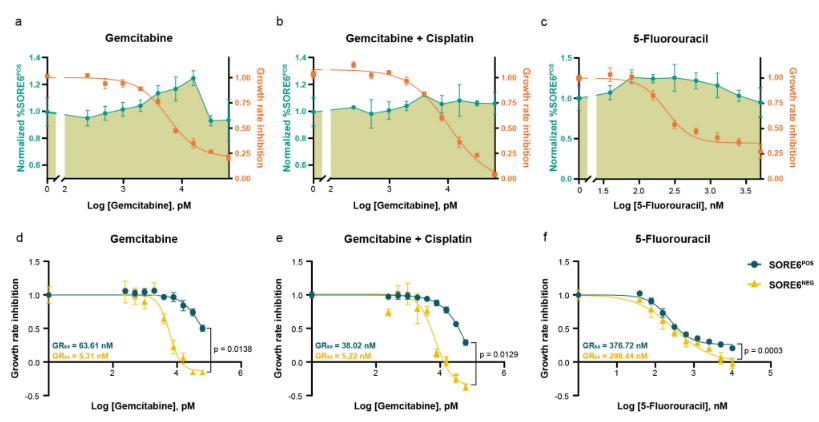
- Algorithmic Drug Comparison: Developed a detailed methodology to create an algorithm for comparing how different drugs affect the CSC proportion across a comparable dose range.
1.3 Methods and Tools:
-
Wet Lab: Cultured CSC reporter-containing cell lines for phenotypic observation and drug response studies.
-
Imaging: Performed high-content and high-throughput imaging, including real-time, live-cell, and fixed-cell techniques.
-
Image Analysis: Conducted single-cell level analysis using Columbus™ software (PerkinElmer).
-
Data Analysis: Utilized RStudio for further data exploration, visualization, and statistical analysis.
3D Cancer Cell Culturing Models
To create a more realistic and complex environment for cancer studies, I developed and investigated advanced 3D culturing models, including both 3D multicellular spheroids and multi-spheroid systems. This approach allowed me to move beyond the limitations of 2D culture and study CSC phenotypes within a more natural, three-dimensional context using high-content imaging.
Methods and Tools:
-
Wet Lab: Created and cultured 3D multicellular spheroids or multi-spheroid systems.
-
Imaging: Performed high-content and high-throughput confocal imaging on live and fixed cells.
-
Image Analysis: Conducted single-spheroid level analysis using Columbus™ software (PerkinElmer).
-
Data Analysis: Utilized RStudio for rigorous analysis to draw meaningful conclusions.
1. Model 3D-1: 3D Multicellular Spheroid
1.1 Analysis overview:
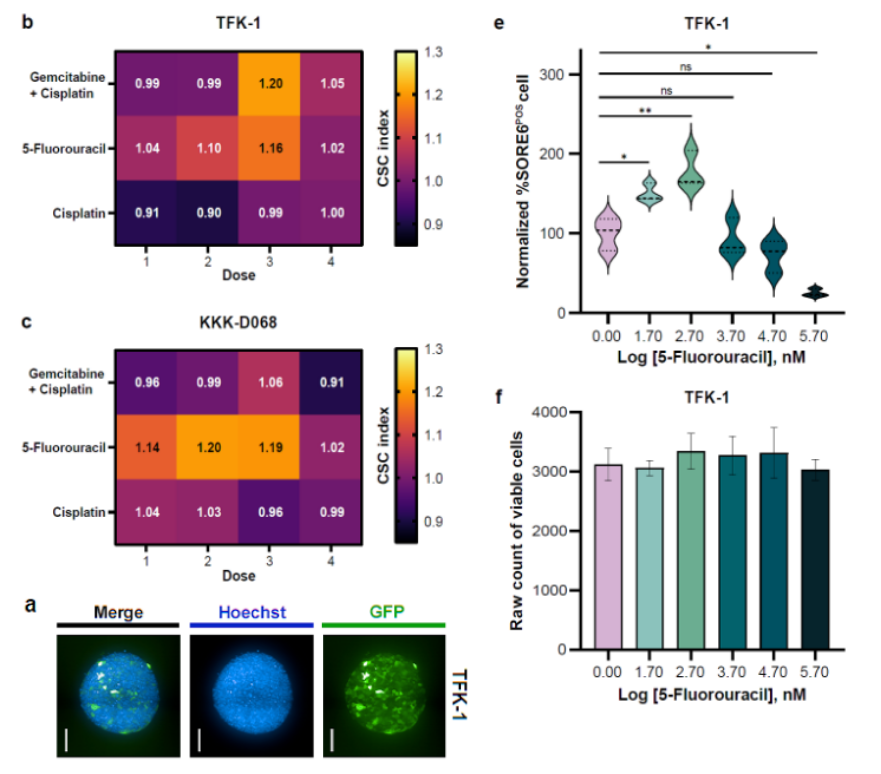
The multicellular spheroid (MCS) model was optimized for cholangiocarcinoma stem cell (CSC) studies. By culturing CCA cells in ultra-low attachment plates, we generated compact spheroids that mimic the tumor microenvironment. High-content confocal imaging enabled precise quantification of spheroid area and CSC signal intensity. Compared with 2D cultures, this model provided a higher CSC yield and revealed drug-induced CSC enrichment in response to chemotherapy. Importantly, the system linked drug concentration to cytotoxicity while simultaneously monitoring CSC dynamics, highlighting its potential in functional precision medicine.
1.2 Key Applications:
- Linking CSC Biology and Drug Response: Established a direct link between a drug’s concentration, its cytotoxic effect, and its parallel impact on the content of CSCs in the 3D Multicellular Spheroid context.
2. Model 3D-2: 3D Multi-Spheroids
We developed 3D-SiSP for 3D multi-spheroid quantitative analysis Project page →
2.1 Analysis overview:
The 3D multi-spheroid model was established to overcome the limitations of large single spheroids, such as necrotic cores and poor imaging penetration. Using methyl cellulose–based suspension, we generated uniform, non-aggregated spheroids across CCA cell lines. We then developed a novel quantitative framework, 3D Surface Integrative Spheroid Profiling (3D-SiSP), which uses area-based measurements from maximum projection images to accurately assess spheroid growth and CSC biosensor signals.
2.2 Key Applications:
- In vitro tumorigenicity: 3D-SiSP allowed more precise measurement of tumor-forming ability than traditional length-based methods. It could detect significant CSC-driven spheroid formation even at very low seeding densities.
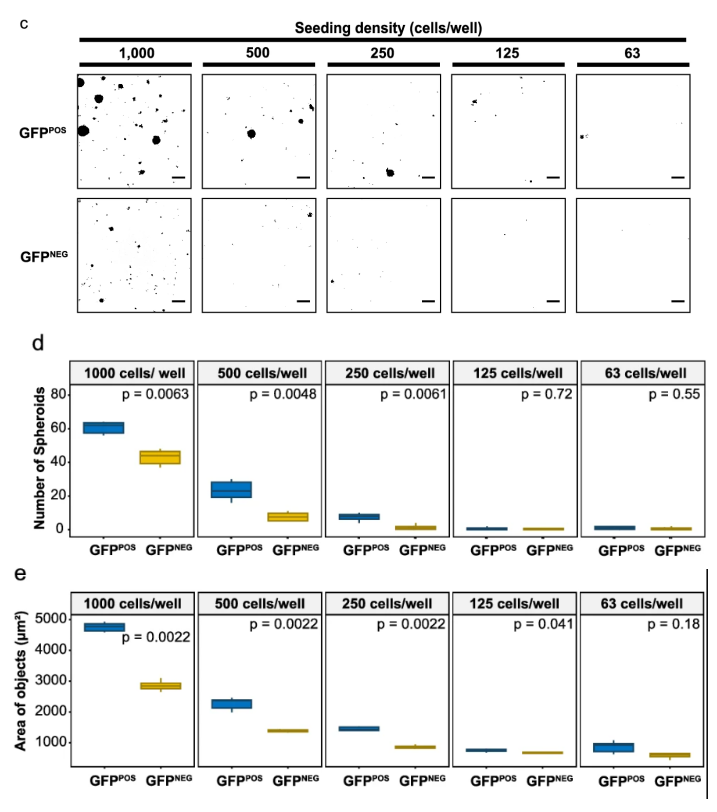
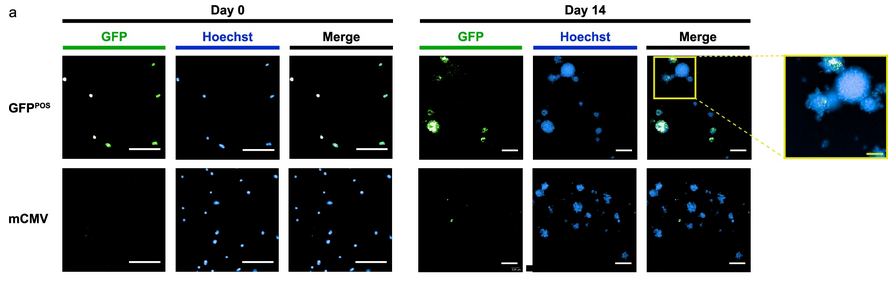
- Size and differentiation: Tracking GFP signals showed that larger spheroids tended to keep more CSCs, while smaller ones often lost stemness and differentiated. This revealed a direct link between spheroid size and cell state.
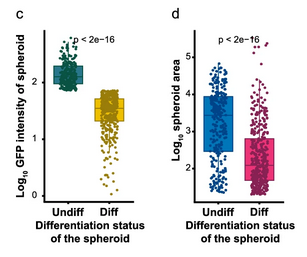
- Drug response: The method quantified both cell killing and CSC enrichment in response to chemotherapy. Interestingly, sublethal doses of 5-fluorouracil and gemcitabine caused paradoxical increases in CSC content.
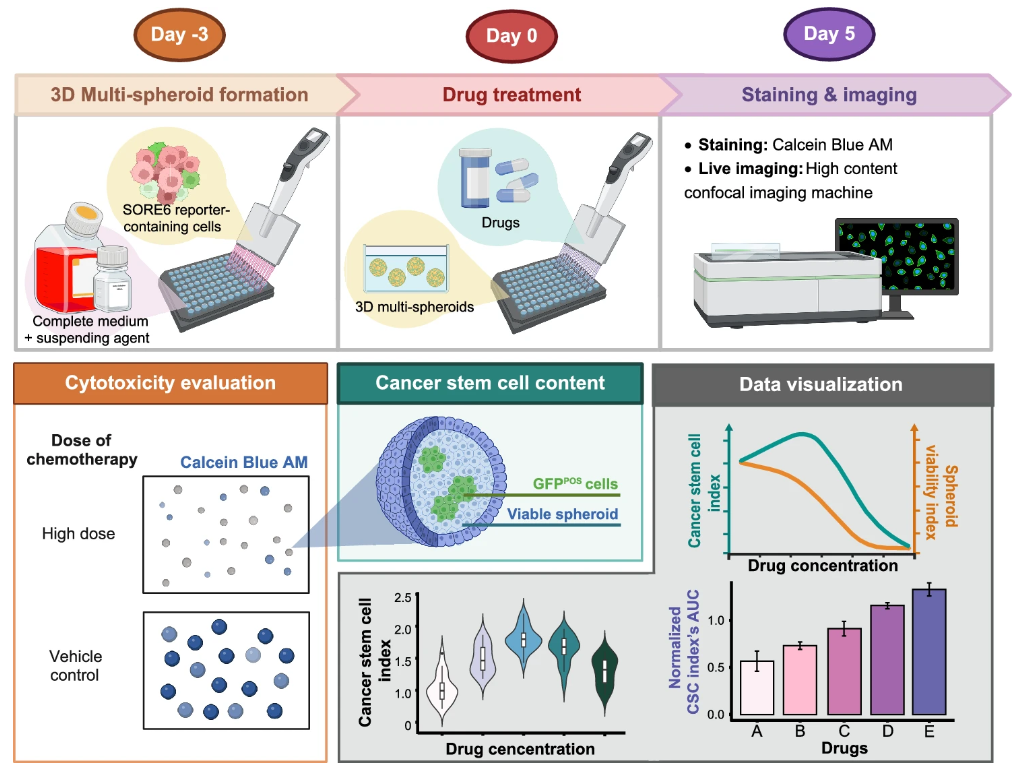
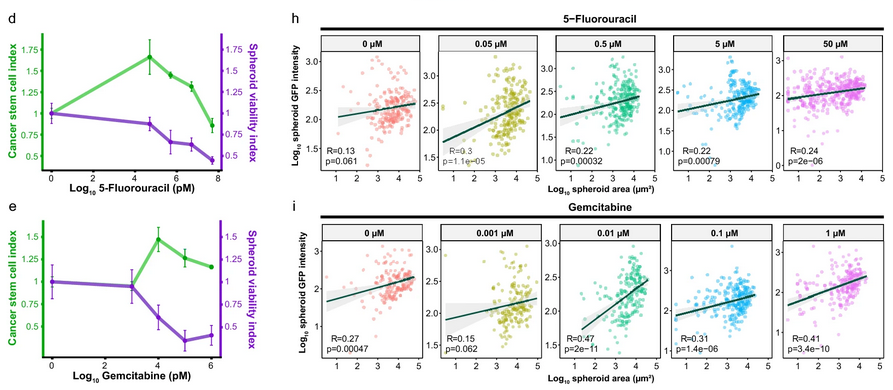
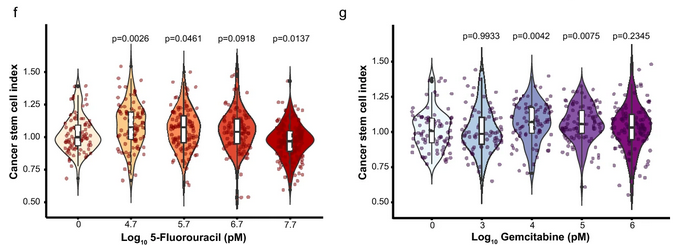
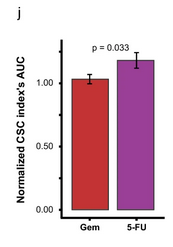
- Drug comparison: By comparing CSC enrichment across different drugs within the same range of spheroid viability, 3D-SiSP highlighted that 5-fluorouracil induced stronger CSC enrichment than gemcitabine.
🔗 Related Projects and Resources
- 3D-SiSP for 3D multi-spheroid quantitative analysis Project page →
- Cholangiocarcinoma Stem Cell ResearchProject page →
What I’ve Learned
There are many ways to approach imaging data, often guided by applications from instrument companies or published studies. However, at its core, we’re really dealing with biology. In my case, it’s the biology of CSCs. I needed to understand the gaps in the CSC field and how I could use available applications to fill them.
I noticed that when studying CSC-mediated drug resistance, researchers often use a single drug concentration to show CSC enrichment or depletion. Yet, in a real tumor, the drug exists in a range of concentrations, not just one. To address this, I created an experiment to analyze the dose-response relationship alongside the CSC content. I was also curious about how different drugs affect CSC content.
Without coding, this kind of single-cell (in 2D) or spheroid (in 3D) analysis would be impossible. Coding speeds up the analysis and enables powerful data visualization. This doesn’t just create beautiful figures; it also provides meaningful insights for exploratory data analysis (EDA).
Acknowledgement
-
I would like to thank my co-authors and colleagues.
🏀 Supawan Jamnongsong, a high-content imaging specialist and a wonderful senior who always provides comments, suggestions, and assistance.
🏀 Pornlada Likasitwatanakul, a talented medical student who I knew could do both wet lab and image analysis from the moment we met.
🏀 It was such a great and exciting time working with you all. I look forward to our next collaboration soon! Haha 😆😆 “Let’s play together!” -
I would like to thank my teachers and my co-advisors.
🚀 Emerita Prof.Dr. Marianne Hokland, who encouraged me to perform 3D culturing experiments.
🚀 Asst.Prof.Dr. Somponnat Sampattavanich, who always provides comments and suggestions on image analysis.
🚀 Asst.Prof.Dr. Methichit Wattanapanitch, who provided intensive and exclusive discussions from a healthy stem cell perspective. -
Lastly, I would like to thank my Ph.D. advisor, Assoc.Prof.Dr. Siwanon Jirawatnotai. Without him, I would not have discovered the exciting world of cancer biology. 🌞🌞 From that point, I expanded my interest to the -omic fields (including Phenomics) for a deeper understanding. 🤩🤩
Last note from me
- I’m sorry for coding resource of 2D model. I will upload the R code to my GitHub as soon as possible. The code is very unreadable (even to me, haha 😆😆!), because these image analysis scripts were some of my very first coding projects. I was practicing by doing real-world data analysis (Yes!, my Ph.D. thesis), and looking back at it now, it feels like I’m recalling a previous life. That’s because of my poor coding practices at the beginning (when I didn’t have any support from an LLM at that time).
I’m very happy 🥰 that you are visiting my computational biology portfolio and would be even happier if you could provide suggestions or feedback 🤩.
You can contact me through various online platforms here 📬 or leave a comment below using GitHub account. 👇🏼